 You can download a model of single triple helix with symmetry information included so you can reconstruct the fibril of the size you want using e.g.Chimera.
You can download a model of single triple helix with symmetry information included so you can reconstruct the fibril of the size you want using e.g.Chimera.
Generate symmetry mates:
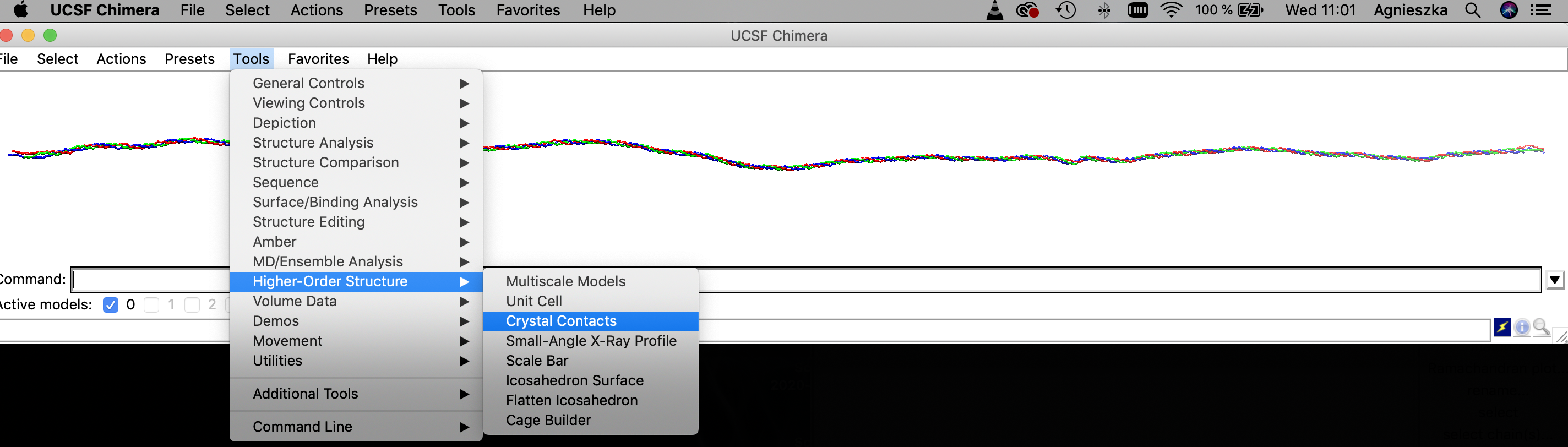
Select contact distance and mark the field "Create copies of contacting molecules
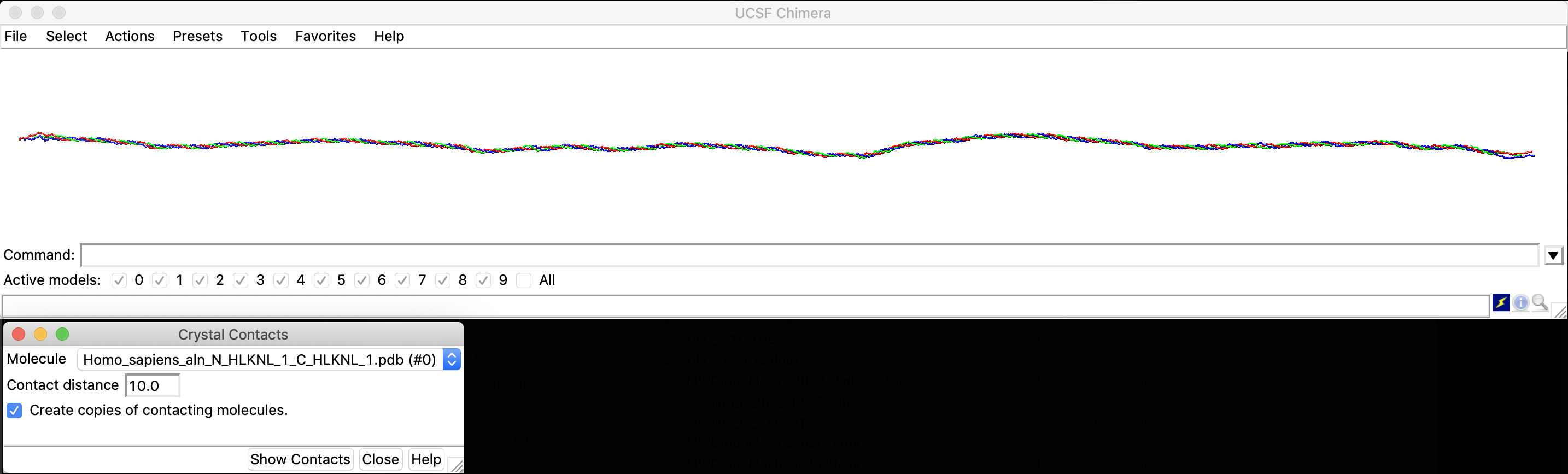
You should see the fibril (here colored by chain id with ribbon's width and height scaled to 1.7)
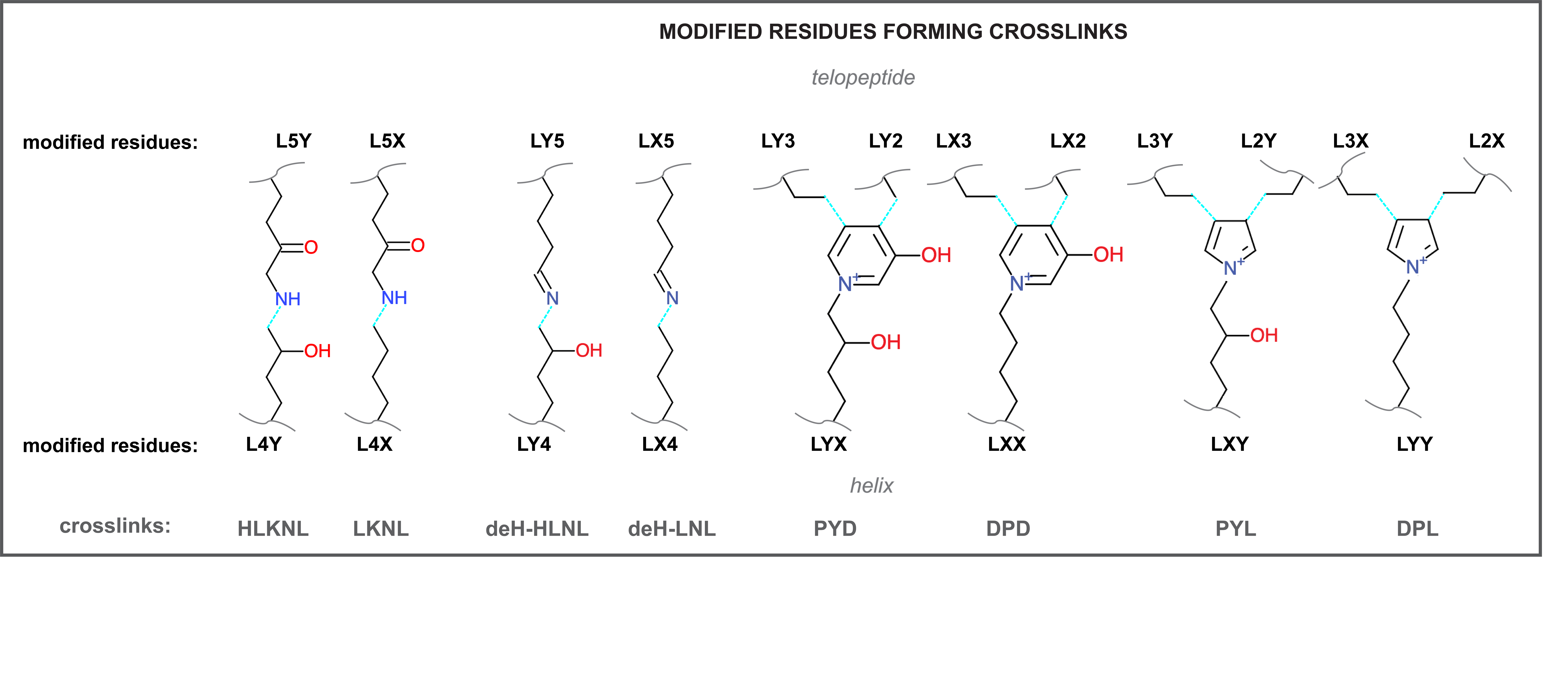
Here are the names of the modified lysine residues forming crosslinks.
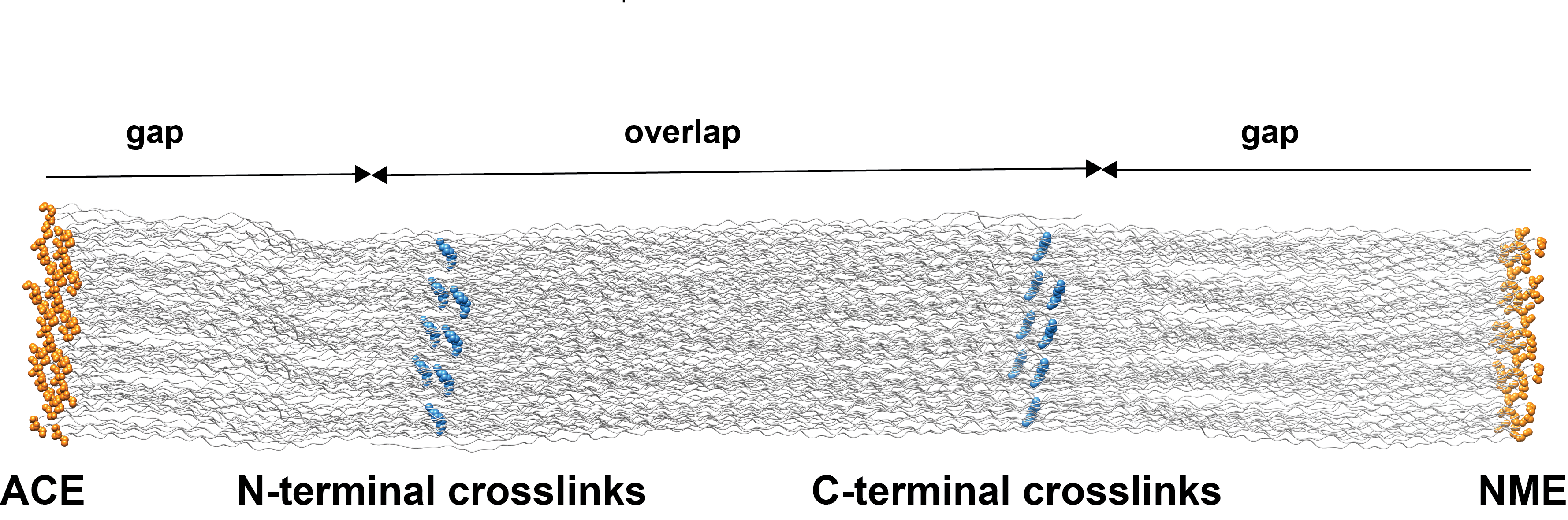
You can also download a model of the fibril. It comprises 41 triple helices spanning one central overlap and one gap region (in two parts, left and right). The caps, ACE and NME are added at the ends of truncated chains (so the models are ready to use in MD simulations).
We suggest to use either Chimera or VMD to visualize a model of the fibril. In Pymol you would encounter problem in ribbon/cartoon representations, because of repetitive chain ids. You can use other representations and command: "set cartoon_gap_cutoff, 0" or contact us. We will provide a script that fixes this issue.
 You can download a model of single triple helix with symmetry information included so you can reconstruct the fibril of the size you want using e.g.Chimera.
You can download a model of single triple helix with symmetry information included so you can reconstruct the fibril of the size you want using e.g.Chimera.



